Learning Update — June 18, 2021
Canals, seaways, cities. More DNA, RNA, proteins.
Canals, Great Lakes, and the St. Lawrence Seaway
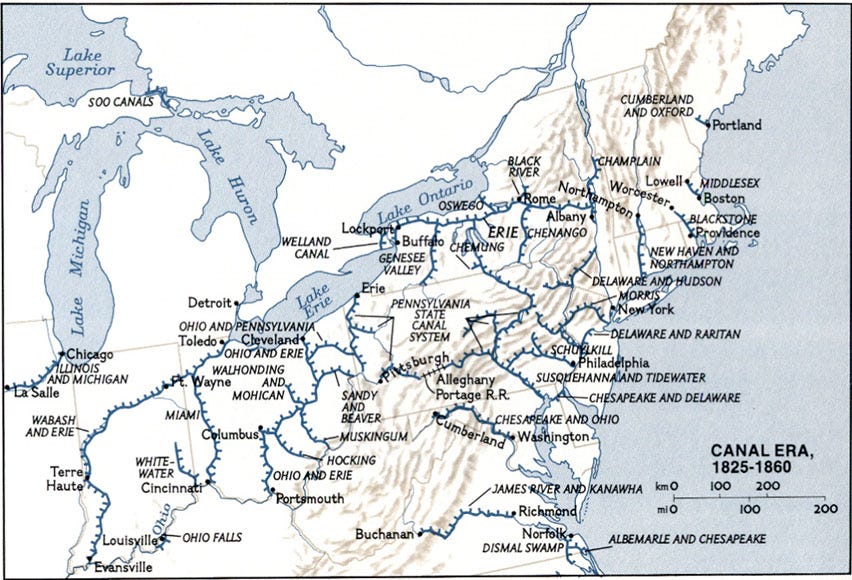
Before there were railroads, there were canals. And like railroads, and then highways, cities and towns that were best connected to the canal network grew and prospered.
The major canals in North America were constructed around 1810-1860. The canals connected inland rivers to major ports; either ocean ports on the east coast (Atlantic), the south coast (Gulf of Mexico), or one of the Great Lakes which were connected to the ocean by the St. Lawrence Seaway.
By the 1850’s, major railroads were already constructed and by the 1900’s, most canals were obsoleted. The major waterways, like the St. Lawrence Seaway, continued to be in use (and are still in use today). Other canals are still in use for nostalgic reasons.
The St. Lawrence Seaway was itself dependent on many canals and locks to bypass rapids, dams, and elevation changes.
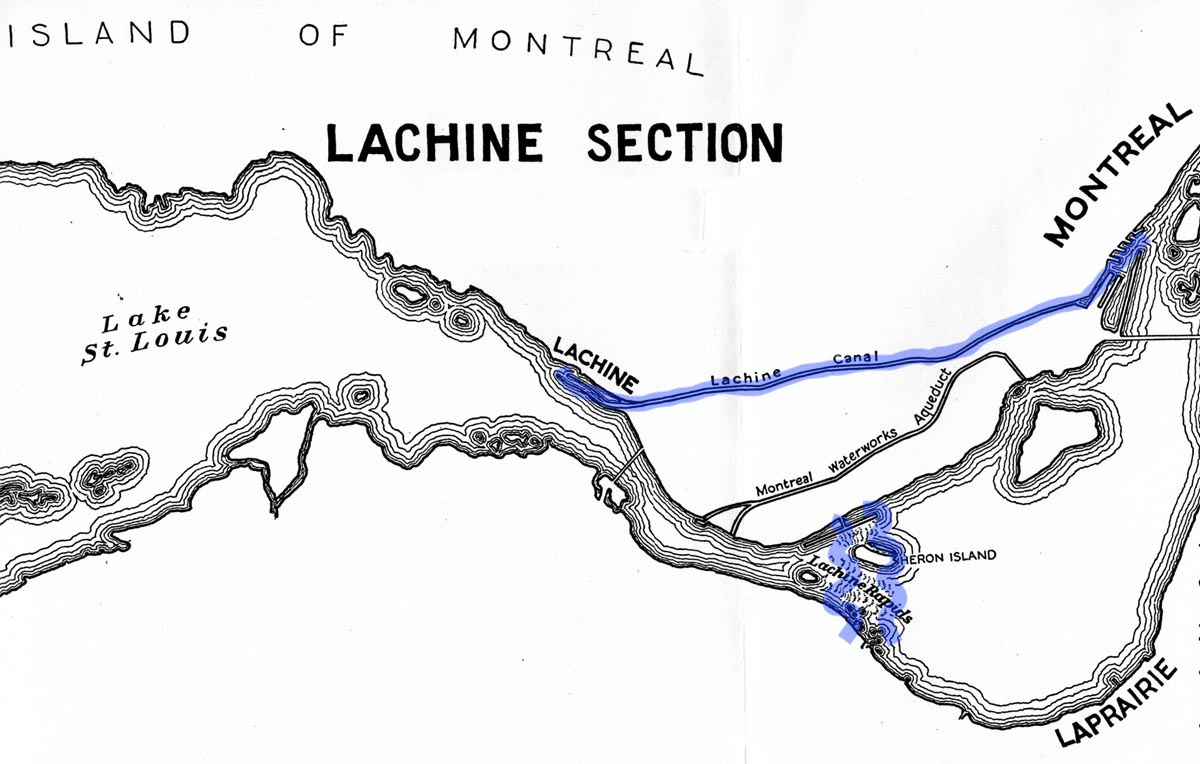
For example, the Lachine Canal in Montreal was constructed to bypass the Lachine Rapids, on the south-west of Montreal Island.
In 1535, on his second voyage to the New World, Jacques Cartier had to turn back at the Lachine Rapids — there was no canal! And the Prairies River on the other side of Montreal Island had even more rapids. The rapids and the inability to sail past them is likely one of the main reasons that the location became a French settlement (Montreal).
Side fact: The rapids obtained the name "La Chine" because Cartier believed the St. Lawrence River was the northwest passage to China and that the rapids were preventing him from getting there.
DNA, RNA, proteins, etc.
The relationship between triplets (codons) and amino acids was worked out by adding various known sequences of mRNA into compounds of burst E. coli cells. The burst cells still contain ribosomes, and all the other materials needed for genetic translation, and will therefore react to the mRNA and manufacture the appropriate proteins.
Here’s how it went down. First, in 1961, Marshall Nirenberg took an mRNA of just U’s and added it to some burst E. coli cells:1
U U U U U U U U U U U U . . .And he ended up, exclusively, with the amino acid phenylalanine.
Then he added an mRNA of just C’s into the burst E. coli cells:
C C C C C C C C C C C C . . .And he ended up, again exclusively, with the amino acid proline.
Then someone else, Har Gobind Khorana, added a chain of UC’s into burst E. coli cells:
U C U C U C U C U C U C . . .And that experiment produced the amino acids serine and leucine.
Now think about it. A simple explanation for this result is that sequences of three nucleosides correspond to an amino acid. In this case, UUU corresponds to phenylalanine, CCC corresponds to proline, while UCU and CUC correspond to one of serine and leucine (although it’s not yet clear which).
But you can figure it out by playing a bit more: Separately, submit (and I am now presumptively grouping nucleotides into groups of three) the sequences:
UCU UUU UCU UUU UCU . . .CUC UUU CUC UUU CUC . . .And we will find that the first sequence produces the amino acids phenylalanine (which we think corresponds to UUU) and serine. And then the second sequence produces the amino acids phenylalanine (again, we think is UUU) and leucine.
Thus we can deduce that UCU specifically corresponds to serine and CUC specifically corresponds to leucine. Etc.
Eventually we get the entire genetic code:
Happy Learning!